import os
import pandas as pd
import numpy as np
import feather
import matplotlib.pyplot as plt
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import PowerTransformer
from sklearn.model_selection import RepeatedStratifiedKFold, GridSearchCV
from sklearn.linear_model import LogisticRegression
from sle.modeling import generate_data, prep_data
from sle.penalization import regularization_range, choose_C, regularization_path, plot_regularization_path, coef_plots_regularized
%load_ext autoreload
%autoreload 2Predict whether an SLE patient has nefritis or not
Citation
This notebook contains analyses for the following project:
Brunekreef TE, Reteig LC, Limper M, Haitjema S, Dias J, Mathsson-Alm L, van Laar JM, Otten HG. Microarray analysis of autoantibodies can identify future Systemic Lupus Erythematosus patients. Human Immunology. 2022 Apr 11. doi:10.1016/j.humimm.2022.03.010
Running the code without the original data
If you want to run the code but don’t have access to the data, run the following instead to generate some synthetic data:
data_all = generate_data('imid')
X_test_df = generate_data('rest')Code for loading original data
data_dir = os.path.join('..', 'data', 'processed')
data_all = feather.read_dataframe(os.path.join(data_dir, 'imid.feather'))
X_test_df = feather.read_dataframe(os.path.join(data_dir,'rest.feather'))cv = RepeatedStratifiedKFold(n_splits=5, n_repeats=5, random_state=40)
trf = PowerTransformer(method='box-cox')y_nefritis = data_all[data_all.Class=='SLE'].Nefritis.astype(np.int64)
X_nefritis = data_all[data_all.Class=='SLE'].drop(["Class"]+["Arthritis","Pleurisy","Pericarditis","Nefritis"] + ["dsDNA1"], axis=1)Xp1_nefritis = X_nefritis + 1 # Some < 0 values > -1. Because negative fluorescence isn't possible, and Box-Cox requires strictly positive values, add ofset
X_trf_nefritis = pd.DataFrame(trf.fit_transform(Xp1_nefritis), index=X_nefritis.index, columns=X_nefritis.columns)clf_lasso = LogisticRegression(penalty='l1', max_iter = 10000, solver = 'liblinear')K = 100
lambda_min, lambda_max = regularization_range(Xp1_nefritis,y_nefritis,trf)
Cs_lasso_nefritis = np.logspace(np.log10(1/lambda_min),np.log10(1/lambda_max), K)
pipe = Pipeline([
('trf', trf),
('clf', clf_lasso)
])
params = [{
"clf__C": Cs_lasso_nefritis
}]
lasso_nefritis = GridSearchCV(pipe, params, cv = cv, scoring = 'roc_auc', refit=choose_C)%%time
lasso_nefritis.fit(Xp1_nefritis,y_nefritis)CPU times: user 4min 53s, sys: 351 ms, total: 4min 53s
Wall time: 5min 49sGridSearchCV(cv=RepeatedStratifiedKFold(n_repeats=5, n_splits=5, random_state=40),
estimator=Pipeline(steps=[('trf',
PowerTransformer(method='box-cox')),
('clf',
LogisticRegression(max_iter=10000,
penalty='l1',
solver='liblinear'))]),
param_grid=[{'clf__C': array([28.71916258, 26.78358714, 24.97846301, 23.2949982 , 21.72499329,
20.26080145, 18.89529124, 17.62181185,...
0.21724993, 0.20260801, 0.18895291, 0.17621812, 0.16434161,
0.15326553, 0.14293595, 0.13330254, 0.1243184 , 0.11593975,
0.1081258 , 0.10083849, 0.09404231, 0.08770417, 0.08179321,
0.07628062, 0.07113956, 0.06634499, 0.06187356, 0.05770349,
0.05381447, 0.05018755, 0.04680508, 0.04365057, 0.04070867,
0.03796504, 0.03540633, 0.03302006, 0.03079462, 0.02871916])}],
refit=<function choose_C at 0x7fb3520bd560>, scoring='roc_auc')Best model:
lasso_nefritis.cv_results_['mean_test_score'].max()0.6029809240238928Score with lambda selected through 1 SE rule:
lasso_nefritis.cv_results_['mean_test_score'][lasso_nefritis.best_index_]0.5939512492715618coefs_lasso_nefritis, nnz_coefs_lasso_nefritis = regularization_path(Cs_lasso_nefritis, clf_lasso, X_trf_nefritis, y_nefritis)ax1, ax2, ax22 = plot_regularization_path(1/Cs_lasso_nefritis, coefs_lasso_nefritis, nnz_coefs_lasso_nefritis, lasso_nefritis.cv_results_)
#ax22.set_ylim([0.8, 1])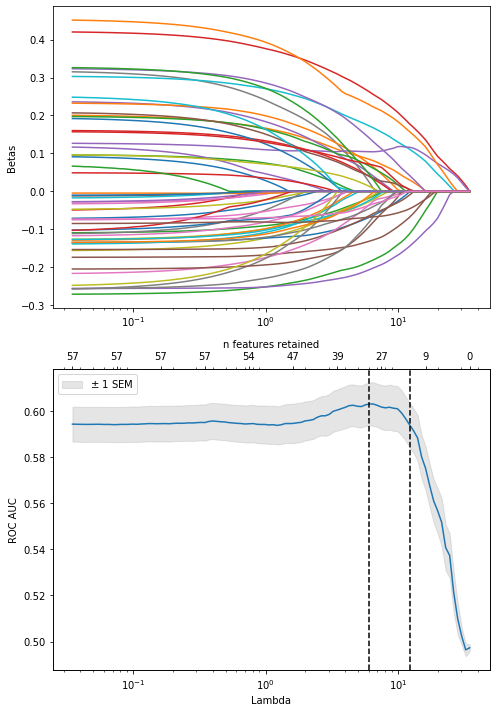
coef_plots_regularized(coefs_lasso_nefritis, nnz_coefs_lasso_nefritis, lasso_nefritis.cv_results_["mean_test_score"], varnames=Xp1_nefritis.columns)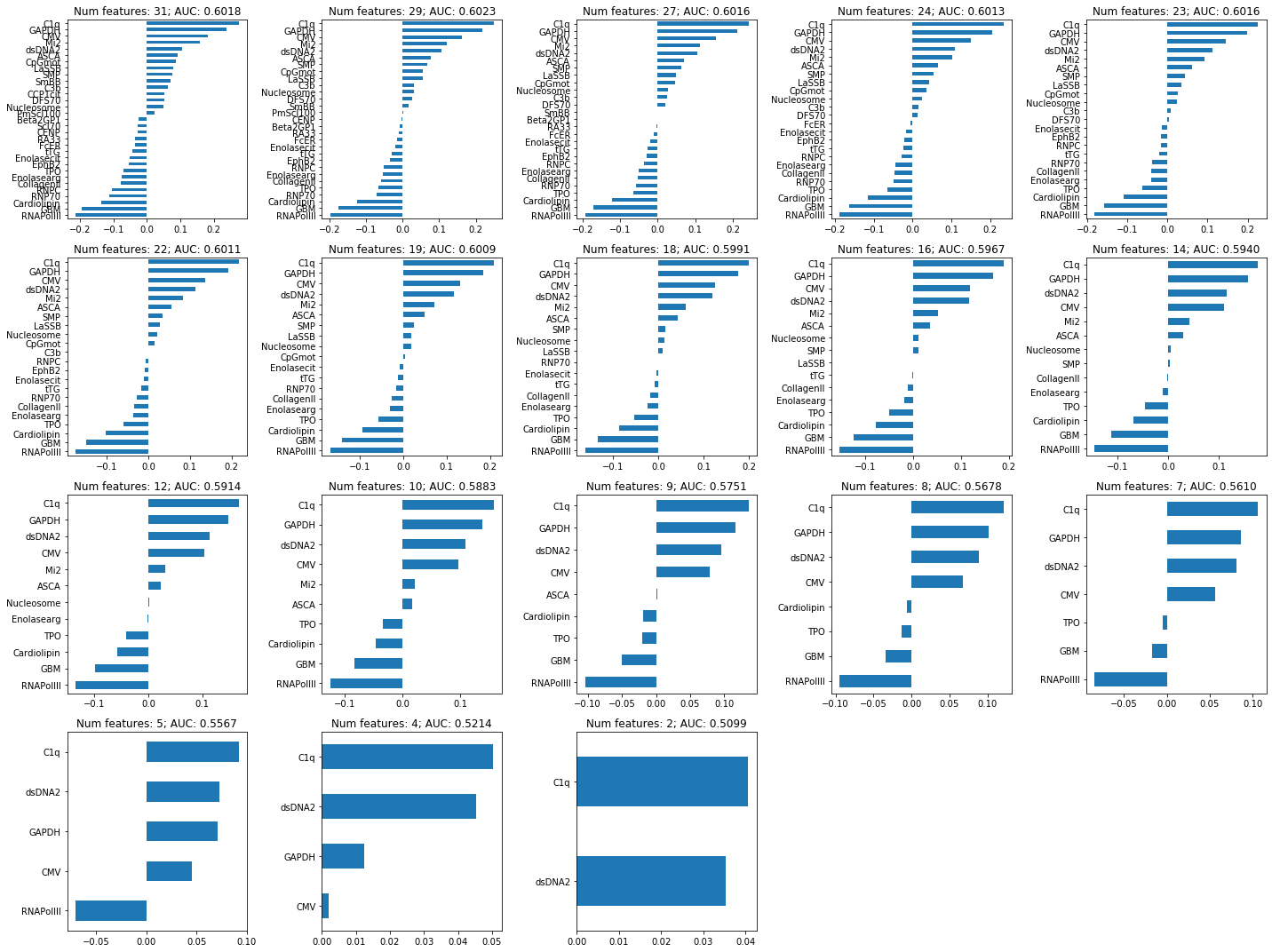
Non-zero coefficients of the model selected with the 1SE rule:
(pd.Series(lasso_nefritis.best_estimator_.named_steps.clf.coef_.squeeze(), index = X_nefritis.columns)[lambda x: x!=0].sort_values(ascending=False))C1q 0.177918
GAPDH 0.157594
dsDNA2 0.116051
CMV 0.110894
Mi2 0.041954
ASCA 0.030033
Nucleosome 0.005160
SMP 0.003712
CollagenII -0.001279
Enolasearg -0.009766
TPO -0.046097
Cardiolipin -0.068313
GBM -0.112145
RNAPolIII -0.144926
dtype: float64